Multimodal Radiomics Platform (MRP)
& Machine-Learning Models
This implementation is aligned with "Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas" (Clinical Cancer Research, 24(18):4429-4436, 2018, doi: 10.1158/1078-0432.CCR-17-3445).
We encourge academic usages of these materials and certainly welcome the test and refinement of the machine-learning models using different populations of glioma (including MR data, information of histology, IDH and 1p/19q statuses)
Developer: Chia-Feng Lu, Ph.D. (alvin4016@ym.edu.tw, +886-2-2826-7308)
Please do fill out the registration form before using any material from this webpage. CLICK HERE
HIGHLIGHTS
Machine learning based on the radiomics of multimodal magnetic resonance images (MRIs) provides an alternative for noninvasive assessment of glioma subtypes in line with 2016 WHO classification. A three-level machine-learning model was proposed to stratify molecular subtypes of gliomas. The accuracies of classifications for the IDH and 1p/19q status of gliomas were between 87.7 and 96.1%. The complete classification of 5 molecular subtypes solely based on the MRI radiomics achieved an 81.8% accuracy, and a higher accuracy of 89.2% could be achieved if the histology diagnosis is available. The proposed approach can assist the molecular diagnosis of gliomas by clinical MRIs.

Multimodal Radiomics Platform, MRP V5.0
Developed by Chia-Feng Lu @ 2018-2022, alvin4016@ym.edu.tw
MRP Latest update: 2022.6.1
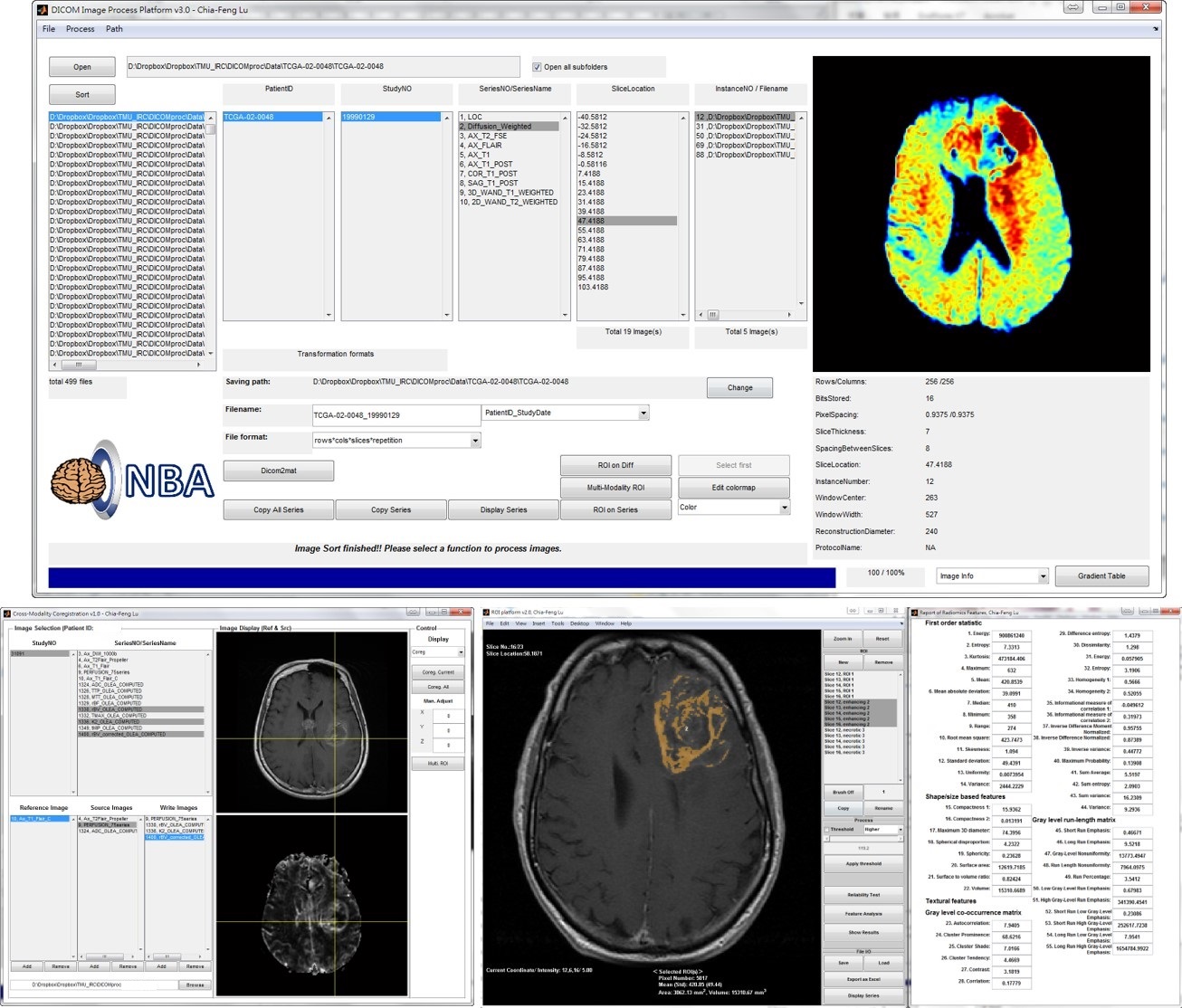
Major Function List of MRP V5.0 (Download Matlab codes)
[Note] Microsoft Excel is required for the output of radiomic features in an xls/xlsx format. Memory larger than 16 GB is recommended.
- DICOM read and sort for MRI, CT, and PET images, optimized for brain, breast, and chest imaging.
- Cross-modality image co-registration and interpolation.
- Multi-modality operation of region of interest (ROI) and thresholding.
- Series images crop, zoom, window/contrast adjustment, and print.
- Computation of Radiomic features, including intensity-based, geometry-based, textural analyses, and wavelet decomposition for each MR modality (CET1, T2W, T2 FLAIR, ADC, Cp/rCBV, Ktrans/K2 maps). The computations on CT and PET SUV maps are also workable.
- Output extracted Radiomic features as Excel sheets.
- [Not released] Correlation and multivariate linear regression analyses between image features and gene expression of tumors.
[Instruction Videos]
- General introduction [01:51]
- Data preparation and DICOM import [09:41]
- Image coregistration and resolution adjustment [10:47]
- Multi-modality operation of region of interest (ROI) and thresholding [19:50]
- Extraction of Radiomic Features (incuding wavelet decomposition) [08:33]
Three-Level Machine-Learning Model in Glioma
Developed by Chia-Feng Lu @ 2017-2018, alvin4016@ym.edu.tw
Architecture of Three-Level Machine-Learning Model (Please see the Figure 1 in the published paper)
- Classification Model for LGG vs. GBM (Download the trained model, threshold=0; Download the MATLAB script to retrain the model).
- Classification Model for IDH status in GBM (Download the trained model, threshold=-0.63; Download the MATLAB script to retrain the model).
- Classification Model for IDH status in LGG (Download the trained model, threshold=1.60; Download the MATLAB script to retrain the model).
- Classification Model for 1p/19q status in IDH-mutant LGG (Download the trained model, threshold=0; Download the MATLAB script to retrain the model).
* This model is pre-trained based on the TCGA-GBM and TCGA-LGG data collection (214 subjects from The Cancer Imaging Archive, http://www.cancerimagingarchive.net/), and is tested on the Taiwanese glioma dataset (30 subjects) and REMBRANDT collection (40 subjects, https://wiki.cancerimagingarchive.net/display/Public/REMBRANDT).